Microbes are fast becoming the darlings of the social behavior set because their interactions can be understood right down to their genes. They do interesting things, too: Bacteria steal iron from each other, kill each other with toxins that only close relatives can resist, and count each other with quorum sensors.
The social amoeba Dictyostelium discoideum, or Dicty for short, is a powerful social study system because of the hard work of generations of cell and molecular biologists who have figured out many of the mechanisms of its social process. But it takes studies in nature to understand whether Dicty’s cooperative behavior benefits relatives, and even whether its social activities occur frequently in nature.
Researchers in the Department of Biology in Arts & Sciences at Washington University in St. Louis are taking a closer look at the wild life of Dicty, using new gene sequencing techniques, as reported in the Proceedings of the National Academy of Sciences.
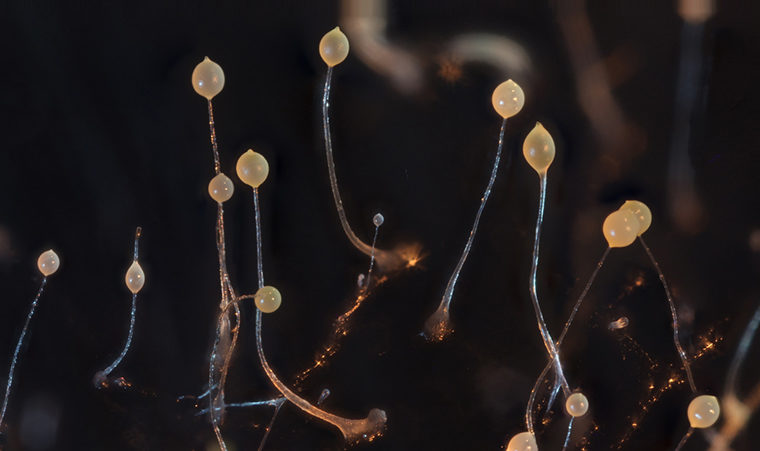
A social study case in point: Triggered by starvation, tens of thousands of formerly independent Dicty amoebae aggregate into a motile slug that ultimately differentiates into a fruiting body with living spores aloft a stalk made of sterile stalk cells. About 20 percent of cells sacrifice themselves to form the stalk that lifts living spores aloft and helps them disperse. This clear separation into altruists (dead stalk cells) and beneficiaries (living spore cells) is reminiscent of an ant colony where the sterile workers assist their queen in reproducing.
But unlike ants, amoebae sometimes form a social unit made up of unrelated individuals. Taking advantage of the sequenced genome and many known genes of Dicty, David Queller and Joan Strassmann and their team — Suegene Noh, Katie Geist and Xiangjun Tian — used a clever technique to get around the difficulty of wild studies of Dicty.

First, they created mixtures, or chimeras, of two unrelated Dicty clones and allowed them to starve. They then collected cells at an early stage when the fate of altruists (stalk cell) and beneficiaries (spore cell) was just beginning to be sorted out, and sequenced their RNA to determine what genes were being expressed in this process. For comparison, they also set up a separate control experiment with only a single clone in the mix. Any genes that were more highly expressed differently in the chimera relative to the control are the social genes of interest. To be sure they were not just looking at the artifacts of one pair of clones, they repeated the experiment with four different clone pairs.

“We started with the prediction that social conflict is going to generate rapid evolution, because every time that you evolve a trick against your opponent, that produces a selection pressure on your opponent to evolve against you, and so on,” said Queller, the Spencer T. Olin Professor of Biology.
The prediction was upheld. Using 15 genomes of Dicty collected by the authors from soils in Virginia and Texas, they found the 78 genes that were expressed differently in chimera were evolving more rapidly than other genes.
“The presence of genes especially activated in chimera and showing rapid evolution is exciting by itself, but also because it supports the validity of laboratory experiments on social interactions,” said Strassmann, the Charles Rebstock Professor of Biology. “The kinds of cheating and competition that we see in the laboratory is thus not an artifact, but something supported by our peek into genes that have evolved over the millennia.”

Said lead author Suegene Noh, now assistant professor at Colby College: “It was particularly exciting to see such a clear signal of rapid evolution from these genes because wild individuals can be highly variable even when they are as apparently simple as single-celled amoebae.”
The researchers also wanted to explore whether cells become stalk or spore based on kin selection or direct selection. The kin selection explanation predicts that stalk cells are altruists towards spore cells that are their kin, while the direct selection explanation is simply that stalk cells are losers in competition. To test this, the investigators looked at genes expressed in the pre-stalk and pre-spore stages. They found that the kin selection predictions were upheld.

“Together, these two experiments demonstrate that, in nature, stalk cells truly are altruists that give up their lives for relatives,” Strassmann said, noting that earlier work had shown that wild fruiting bodies are largely clonal. “But that does not negate the finding that when there are non-relatives in the fruiting body, genes turn on that are under competing selection pressures.
“It’s just like ‘Alice in Wonderland,’ who discovered that she had to run constantly to merely stay in the same place,” she said.
This study takes advantage of molecular signatures of evolution in ways that are likely to prove useful in other controversies of microbial social interactions, the authors said.
“All it takes is a question, a target set of genes hypothesized to be under a particular kind of selection, and then a measure of the genetic signature of that selection,” Queller said. “For this particular system, it validates the laboratory work that has been going on for decades, an advantage that could also come to other systems.”