
Within both plant and animal cells, motor proteins act like the engines in a busy train system. They shuttle material in the cell from one location to another. And just as commuter trains travel a predictable route in a defined direction, their volume of transport is commensurate with need. At rush hour, more trains are in operation. At midnight, there’s no point in running trains every 10 minutes.
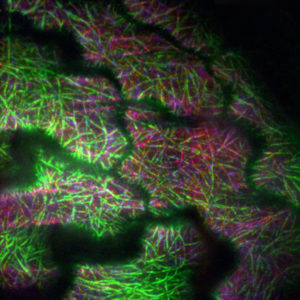
In a growing plant cell, motor proteins called kinesins work as transporters that haul materials built in one part of the cell to the place where they are needed. Kinesins travel along tracks of polymers known as microtubules to get where they are going. Moving cargo costs the cell energy and resources, and this process is closely controlled to prevent waste.
Now biologists at Washington University in St. Louis have discovered the molecular brakeman that holds kinesins in check until their cargo is needed.

The importin IMB4 is a regulator that controls a kinesin specifically involved with building the plant cell wall. It works by physically binding to the kinesin, said Ram Dixit, associate professor of biology in Arts & Sciences. IMB4 holds the kinesin in an inactive state — protecting it from degrading while it waits — and prevents the kinesin from traveling along a microtubule until its cargo is needed. The new research is published in the journal Developmental Cell.
“The cell wall is like the plant’s exoskeleton, and building it is one of the growing plant’s most important functions. We have identified a key molecular regulator that closely controls cell wall deposition by physically binding to a kinesin,” Dixit said. “We still don’t know what signals cause IMB4 to release the brakes, but we now understand how it holds kinesins back until they are needed.”
Ready movers
A rigid cell wall is an essential and energetically expensive investment for a plant. The cell wall confers strength and enables the cell to withstand the turgor pressure that is necessary for growth.
In Arabidopsis plants, cell wall deposition depends on a kinesin-4, called Fragile Fiber 1 or FRA1. FRA1 was identified more than a decade ago, but a 2015 study in Dixit’s lab confirmed that its motility and abundance were correlated with cellular growth rates.
Postdoctoral fellow Anindya Ganguly, lead author of the paper, was intrigued by what the research team had seen. In rapidly growing cells of seedling stems, they saw lots of FRA1 kinesins moving along microtubules. But further down the stem, where the cells had ceased to elongate, the motor proteins were gone.
“Similar to rush hours, when plants are rapidly growing, you need to deliver a lot of cell wall material to keep up with growth,” Ganguly said. “Efficient, high-volume transport depends on having a lot of motor proteins. The regulator we discovered keeps a surplus of these transporter proteins around during times of rapid growth.”

Ganguly and co-authors discovered that the importin IMB4 binds directly to the motor domain of the FRA1 kinesin. Then, using a combination of mutant analysis, microscopy of FRA1 within living cells and protein biochemistry, the team showed that this interaction inhibits the movement of FRA1.
“This importin essentially jams the motor, the engine of the kinesin,” Dixit said. “You inhibit mobility by virtue of prohibiting its association with its track.”
“The site where we think it binds, based on some analysis we did here, includes amino acids that are very conserved in other kinesin families in both plants and animals,” Dixit said. “So we think that there’s a good chance that this mechanism might be generally applicable.”
Twisted sister
With a handle on IMB4, researchers now have a better understanding of the mechanical workings of the engines of cell wall deposition. Another study published by the Dixit lab this month identifies a regulator involved in building the scaffold on which kinesins move.
In this study, the researchers examined a mutant Arabidopsis plant that exhibits a twisted growth pattern. Its leaves and stems swirl back in on themselves; its roots are tangled and skewed.
These so-called spiral mutants suffer from misaligned microtubules (the “tracks” that the kinesins walked along in the other study). In the mutant, severing proteins were thought to snip and shorten the microtubules at the points where they cross over each other.

But when Dixit and postdoctoral fellows Yuanwei Fan and Graham Burkart set out to parse intracellular activity in the spiral mutant, they uncovered a more interesting and nuanced mechanism. Importantly, they discovered that the plant-specific SPR2 protein regulates the minus-end dynamics of microtubules in the mutant.
Microtubules have a fast-growing and more dynamic end, identified as a “plus” end, and a slow-growing, less dynamic “minus” end. Microtubules grow and shrink from the plus side, while the minus side has generally been considered static in plants — until now.
Remarkably, in the spiral2 mutant, the minus-ends are extremely dynamic and shorten at much higher rates compared to wild-type plants.

“Not only did we find that this protein localizes to the minus end, in plants, and regulates the minus-end dynamics, but we were able to recapitulate that behavior in vitro,” Dixit said. “We can say that this protein and this protein alone is sufficient to localize to the minus ends, track the minus ends, and stabilize them.”
Interestingly, the SPR2 plant protein shares some characteristics with mammalian proteins that stabilize the minus ends of microtubules in the tissues that line the surfaces of organs and in neurons.
This separate, microtubules work is funded by the National Institutes of Health, and is published online in advance of the March 19 issue of the journal Current Biology.
Keeping track of cell shapes and functions
Taken together, this new research on the regulators of motor proteins and microtubules helps explain the inner workings of plant cells that gives rise to their shape and enables them to alter their growth in response to developmental and environmental signals.
The work on IMB4 and the FRA1 kinesin is part of Dixit’s effort with the Center for Engineering MechanoBiology (CEMB), a National Science Foundation-funded science and technology center jointly led by Washington University and the University of Pennsylvania. Within CEMB, Dixit leads a research cohort focused on determining how cells adapt to, and change, their mechanical environment.
“The center’s focus is really to uncover, understand and ultimately be able to control the role of mechanical forces writ large on biological systems,” Dixit said.
“One of the thing that’s unique about the center is that it brings together both plant and animal systems. That hasn’t been done before,” Dixit said. “The idea is that we will be able to uncover some universal principles that are applicable to both kingdoms.”



