Heme is an essential part of the protein hemoglobin, which colors human blood red. Heme also is crucial for cytochrome proteins, which power the cell. Humans, animals, plants and bacteria all use heme.
Hemoglobin shuttles oxygen to tissues where it is needed, while cytochromes carry electrons for energy conversion in the cell. But understanding how heme moves across membranes — like it needs to, in order to insert into hemoglobin and cytochromes — has been challenging. Heme transport is transient, which means heme moves through membranes quickly and leaves behind no traces. And heme-binding membrane proteins are difficult to purify in large quantities.
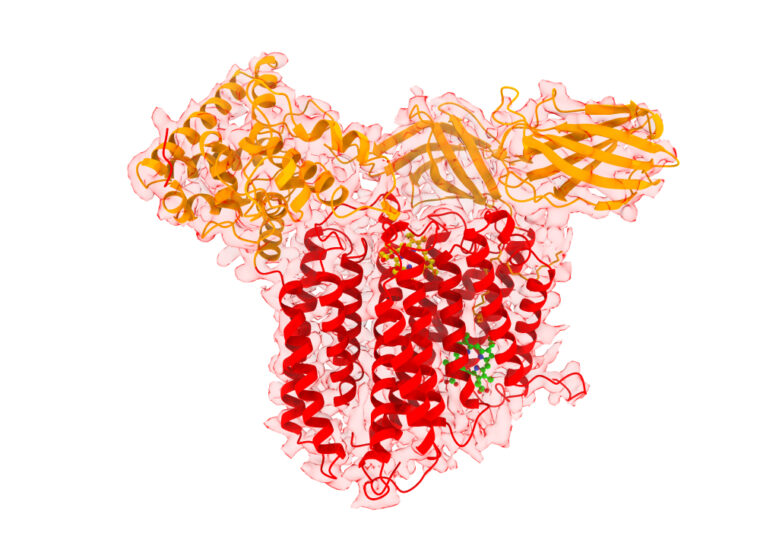
In research published Dec. 20 in Nature Chemical Biology, scientists at Washington University in St. Louis described for the first time the structure of a bifunctional protein, called CcsBA, that transports heme and attaches it to cytochromes. The study led by Robert Kranz, professor of biology in Arts & Sciences, captured two conformational states of CcsBA, a bacterial and chloroplast protein, allowing scientists to characterize the enzyme mechanism.
“This new paper addresses the structural basis for how the CcsBA machine functions, revealing major dynamic switches that occur during the cycle of heme transport,” Kranz said.
The study was made possible by a collaboration with James Fitzpatrick, director of the Washington University Center for Cellular Imaging (WUCCI) at the School of Medicine and a professor of neuroscience, of cell biology and physiology, and of biomedical engineering, and Michael Rau, a staff scientist and structural biologist on his team. They leveraged a cutting-edge structural biology technique called single particle averaging, which utilized a state-of-the-art cryo-Electron Microscope (cryo-EM) to image different views of the protein in its natively vitrified (frozen) state. After sorting all of the different views, they were able to construct cryo-EM density maps — which are three-dimensional representations of the protein built from a series of two-dimensional projections of various views — from which Kranz’s team could build an atomic model of the structure of CcsBA.
“Cryo-EM is a transformative technology that allows us to visualize at the near-atomic level the structural arrangement of a given protein, including the ability to tease out different conformations from an ensemble of states,” Fitzpatrick said. “It was this latter ability that was key in enabling us to capture the mechanism of heme transport.”
The cyro-EM data identified two states in which either one or two heme molecules were bound.
“The structural models we were able to construct illustrate that CcsBA is trapped with heme in two different conformations, which we term the closed and open states,” Kranz said. “This new body of work addresses the structural basis by which the CcsBA machine functions, revealing a major dynamic switch that occurs during the transport cycle.
“One of the coolest findings is that a large chamber opens upon heme transport,” he said. The chamber is for cytochrome c synthesis.
Co-first author Deanna L. Mendez, a postdoctoral staff scientist in biology, previously co-authored a study with Kranz in eLIFE about the reconstitution of the purified bacterial and human synthases of heme. CcsBA is different from the human form of heme-transporter/cytochrome c synthase. The insights gained through identifying its structures give researchers a leg up on developing antimicrobial agents that will selectively target bacteria.

“In CcsBA, we observe a clear path between transmembrane alpha helices that could allow heme to travel from the transmembrane-heme site to the external heme site,” Mendez said. “Since heme goes down its concentration gradient during export, we do not envision an energy source requirement for this process.”
Co-first author Ethan Lowder is a senior at Washington University and worked on this project for two years.
“Solving a novel structure of a protein is very difficult,” Lowder said. “In this case, there were no similar structures that we could rely on as a template or starting point. It was definitely a challenge, but we worked all the way through it to get to the final structures.”
The second author, Dustin Tillman, who was an undergraduate at Washington University when he completed this work, was instrumental in purifying CcsBA. He partnered with the WUCCI team to optimize the sample preparation for the single particle cryo-EM studies. “Dustin also performed reconstitution assays on his purified preps to show they were active synthases,” Kranz said.
“I am grateful for the contributions and involvement of our talented undergraduate researchers in this NIH-supported research,” Kranz said. “This study is a culmination of over three decades that our lab has studied heme transport and cytochrome assembly, so it is satisfying to know the structural basis for both. It will lead to more experiments on mechanisms of chamber opening, transport and the synthase reaction in the chamber.”
Funding: This work was funded by the National Institutes of Health, National Institute of General Medical Sciences (R01 GM47909 to Robert G. Kranz). James Fitzpatrick is supported by the Chan Zuckerberg Initiative as an Imaging Scientist (2020-225726) and the Washington University Center for Cellular Imaging (WUCCI) is supported by Washington University School of Medicine, The Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (CDI-CORE-2015-505 and CDI-CORE-2019-813), the Foundation for Barnes-Jewish Hospital (3770) and the Washington University Diabetes Research Center (NIH P30 DK020579).