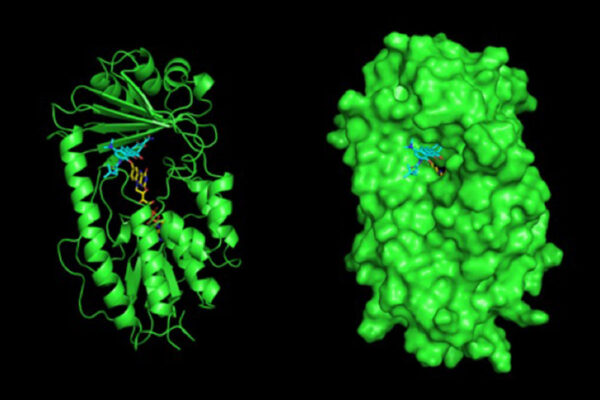
Understanding how antibiotic scaffolds are constructed in nature can help scientists prospect for new classes of antibiotics through DNA sequencing and genome mining. Researchers have used this knowledge to help solve the X-ray crystal structure of the enzyme that makes obafluorin — a broad spectrum antibiotic agent made by a fluorescent strain of soil bacteria. The new work from Washington University in St. Louis and the University at Buffalo was published July 31 in the journal Nature Communications.
A multi-part enzyme called a nonribosomal peptide synthetase produces the highly reactive beta-lactone ring that is responsible for obafluorin’s antimicrobial activity.

“Obafluorin has a novel structure compared to all FDA-approved antibiotics,” said Timothy Wencewicz, assistant professor of chemistry in Arts & Sciences. “In the long term, we really need new structural classes of antibiotics that have never been contaminated by clinical resistance from established antibiotic classes.”
These chemicals could be used as next-generation antibiotics for humans, or even to benefit the agriculture sector, Wencewicz noted — as researchers strive to engineer seed treatments and biopesticides to support plant systems capable of making enough food to feed the 9.6 billion people projected to live on this planet by 2050.
The new work provides a useful road map that shows how individual protein domains in the ObiF1 enzyme are stitched together in three-dimensional space. An enzyme’s structure is fundamental to almost every function it performs.
“The solution of this structure expands on previous discoveries to provide views of the molecular interactions between catalytic domains in a brand new way,” said Andrew M. Gulick, associate professor in the department of structural biology in the Jacobs School of Medicine and Biomedical Sciences at the University at Buffalo. “This is a brand new class of compounds, and we’ve never had the molecular vision to appreciate how they are produced.”
Pinning down a new antibiotic from nature
Obafluorin is made by a fluorescent strain of soil bacteria that forms biofilms on plant roots. Obafluorin was originally discovered in 1984, but it wasn’t until 2017 that Wencewicz uncovered the genetic blueprint of the enzyme that makes the molecule’s bio-active components. That discovery marked the first time that anyone had been able to pin down a beta-lactone forming enzyme from nature, and recreate it in the laboratory.
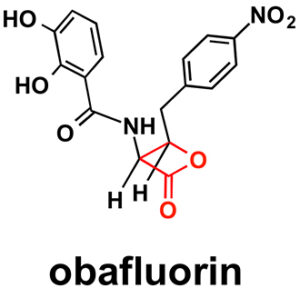 Like penicillin, obafluorin has a four-membered ring — sometimes called an enchanted ring. A four-membered ring puts strain on bond angles that carbon prefers to adopt. But because a four-member ring is unstable, these molecules are also short-lived and difficult to make. For example, it took years for chemists to learn how to synthesize penicillin from chemicals and then figure out how fungi make it. This ultimately led to the global production of penicillin by fermentation.
Like penicillin, obafluorin has a four-membered ring — sometimes called an enchanted ring. A four-membered ring puts strain on bond angles that carbon prefers to adopt. But because a four-member ring is unstable, these molecules are also short-lived and difficult to make. For example, it took years for chemists to learn how to synthesize penicillin from chemicals and then figure out how fungi make it. This ultimately led to the global production of penicillin by fermentation.
Researchers in the Wencewicz laboratory were able to fast-track the discovery process using genetics to zero in on the biosynthetic machinery that bacteria use to make obafluorin, and then to reconstruct that multi-step, enzyme-catalyzed process in the laboratory.
Captured ‘in the act’
For their new work, Wencewicz and Gulick, along with postdoctoral researcher Dale F. Kreitler (UB) and graduate students Jason E. Schaffer and Erin M. Gemmell (Washington University), mapped the full-length nonribosomal peptide synthetase (NRPS) that makes the bio-active components of obafluorin.
Gulick is an expert in NRPS systems and has published many of the seminal crystal structures of core pieces of these enzymes, as well as structures for full-length systems such as the one presented in this work.
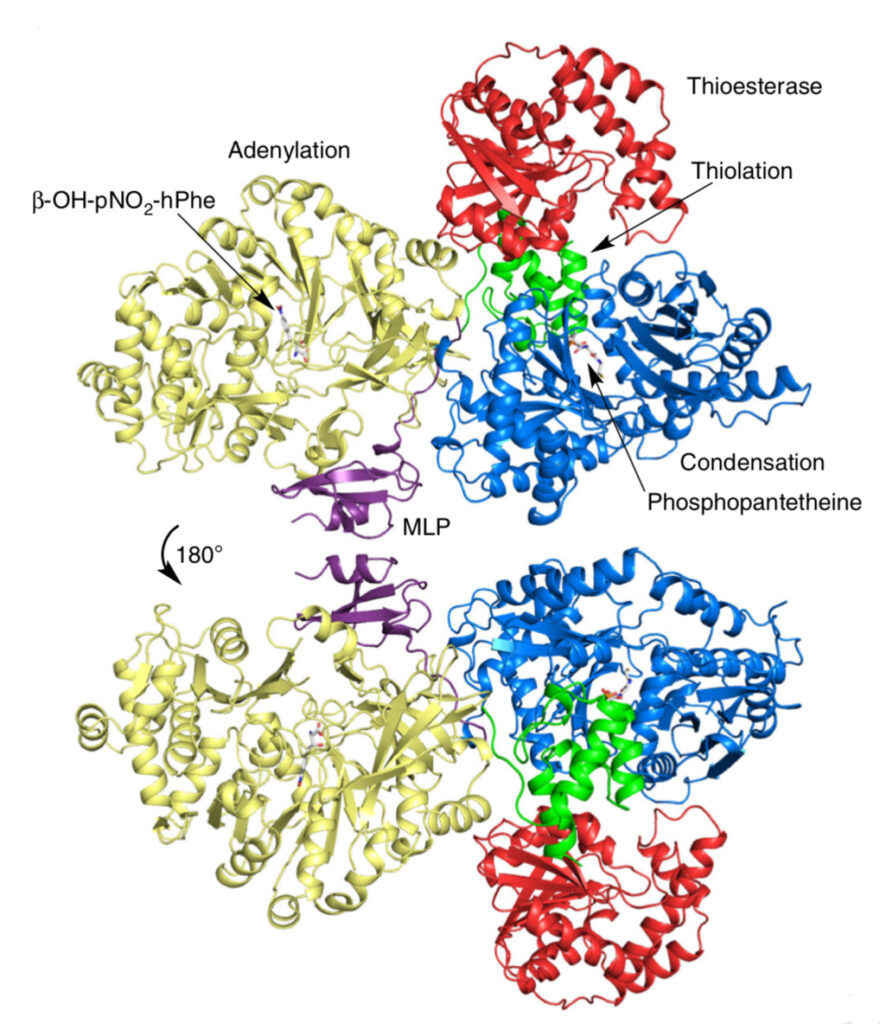
The result is a comprehensive, detailed molecular structure at 3 Angstrom resolution that allows one to identify the atoms in the protein chain, see their location and points of contact along the chain, and determine how the pieces are assembled to produce useful molecules from start to finish.
“We were able to catch some of the building blocks of the molecules captured inside some of the enzyme-active sites — in the act of doing the chemistry,” Wencewicz said. “This helped us to connect small molecules to the protein, and fill in some of the mechanistic gaps in how the molecules are created.”
This type of view is particularly important for the future goal of creating a chemical library, populated with related beta-lactone compounds that have been engineered for beneficial uses.
“We also got a very interesting glimpse at how the domains of the protein actually talk to each other,” Wencewicz said. The crystal structures allowed them to see how key components of the enzyme dock to each other, and how molecules are efficiently moved through the entire NRPS assembly line.
One particular component — something called an MbtH-like protein, or MLP, because it was first identified in a related system to produce mycobactin in the bacteria that causes tuberculosis — was shown to play a critical role in facilitating protein-to-protein interactions between catalytic domains.
“Turns out, the (overall) protein suffers largely when you take away this very little protein that provides the critical handshake,” he said. “It showed us once and for all that the coordination of these domains is absolutely critical to the function of making antibiotics with this enzyme.”
Taken as a whole, the new paper is unique in its reach, the researchers said, presenting the X-ray crystal structure of the complete obafluorin-synthesizing enzyme ObiF1, probing the molecular mechanism for various key steps in the catalytic cycle, and establishing the conserved residues that enable formation of the reactive beta-lactone ring.
A key step in a long process
Finding an antibiotic from a source in nature is only a first step in a long process of drug development. With the structure and techniques reported in this new paper, it is now possible to quickly and easily create analogs of the natural product in the laboratory — to optimize its molecular properties and bioactivity.
Gulick and Wencewicz plan to continue to apply their knowledge of the obafluorin biosynthetic pathway to discover new beta-lactone natural products using genomic and biochemical approaches.
“Given the structural diversity of known beta-lactone natural products, we believe that novel beta-lactone synthases remain to be discovered,” Wencewicz said.
Washington University has filed for a U.S. utility patent covering this technology.